Chapter 10 Synthesis
10.1 Summary
In this session, we’ll pull together the skills that we’ve learned so far. We’ll create a new GitHub repo and R project, wrangle and visualize data from spreadsheets in R Markdown, communicate between RStudio (locally) and GitHub (remotely) to keep our updates safe, then share our outputs in a nicely formatted GitHub ReadMe. And we’ll learn a few new things along the way!
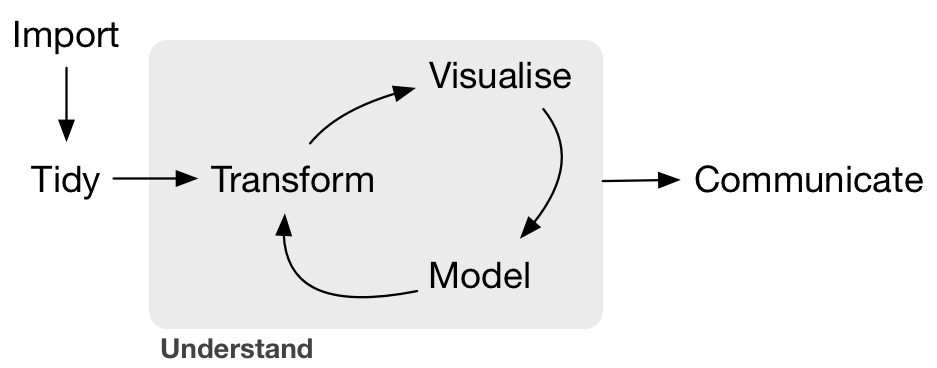
Grolemund & Wickham R4DS Illustration
10.2 Objectives
- Create a new repo on GitHub
- Start a new R project, connected to the repo
- Create a new R Markdown document
- Attach necessary packages (
googlesheets4,tidyverse,here) - Use
here::here()for simpler (and safer) file paths - Read in data from a Google sheet with the
googlesheets4package in R - Basic data wrangling (
dplyr,tidyr, etc.) - Data visualization (
ggplot2) - Publish with a useful ReadMe to share
10.3 Resources
- The
herepackage - googlesheets4 information
- Project oriented workflows by Jenny Bryan
10.4 Lesson
10.4.1 Set-up:
- Log in to your GitHub account and create a new repository called
sea-creature-synthesis - Clone the repo to create a version controlled project (remember, copy & paste the URL from the GitHub Clone / Download)
- In the local project folder, create a subfolder called ‘data’
- Copy and paste the
fish_counts_curated.csvandlobster_counts.csvinto the ‘data’ subfolder - Create a new R Markdown document within your
sea-creature-synthesisproject - Knit your .Rmd to html, saving as
sb_sea_creatures.Rmd
10.4.2 Attach packages and read in the data
Attach (load) packages with library():
Now we’ll read in our files with readr::read_csv(), but our files aren’t in our project root. They’re in the data subfolder.
Use here::here() to direct R where to look for files, if they’re not in the project root. Not sure where that is? Type here() in the Console, and it will tell you!
"/returns/your/project/root/"
Go ahead, find your project root!
Then use here::here() again to easily locate a file somewhere outside of the exact project root. In our case, the files we want to read in are in the data subfolder - so we have to tell R how to get there from the root:
# Read in CSV files
fish_counts <- readr::read_csv(here::here("data", "fish_counts_curated.csv"))
lobster_counts <- readr::read_csv(here::here("data", "lobster_counts.csv"))Check out the two data frames (fish_counts and lobster_counts).
The fish_counts data frame is in pretty good shape. But the lobster_counts df could use some love, because there are “-99999” entries indicating NA values, and the column names would be difficult to write code with.
When reading in the lobster data, let’s:
- convert every “-99999” to an
NA - get the column names into lower snake case using
janitor::clean_names()
lobster_counts <- read_csv(here::here("curation", "lobster_counts.csv"),
na = "-99999") %>%
clean_names() Look at it again to check (always look at your data) - now both data frames seem pretty coder-friendly to work with.
10.4.3 Data wrangling
- join?
- filter?
unite/separate
- Read in lobster data
- Join with another existing data frame (or 2?)
- Pivoting
- Transforming / subsetting
- Grouping & summarizing (for means, sd, count)
- Make a table
Make a graph
Possible new things: complete()